ISBI 2017
Publication in ISBI 2017 Nuclei Segmentation in Histopathology Images Using Deep Neural Networks.
At the ISBI 2017 held in Melbourne, Australia I presented our conference paper Nuclei Segmentation in Histopathology Images Using Deep Neural Networks. The slides for the talk can be found here and paper.
Paper
The conference paper Nuclei Segmentation in Histopathology Images Using Deep Neural Networks by Peter Naylor, Marick Laé, Fabien Reyal and Thomas Walter has been published in Biomedical Imaging (ISBI 2017), 2017 IEEE 14th International Symposium on (pp. 933-936).
Data publicaly available
The data used for the study can be found here. The files come as a zip file and has many subdirectories. Patients histopathological data can be found under “Slide_id” and the ground truth can be found under “GT_id” where id is the patients id. Histopathology data are under a standard RGB files, however the ground truth is under an itk-snap format which is nii.gz. One could use the nibabel to open such images.
Summary of the paper
We note 3 contributions:
- In this paper we introduce our new nuclei detection dataset within HE stained histopathology images. In this dataset, each patient has 3 to 7 annotated 512x512 samples from their original WSI. It is a pixel wise annotation, i.e. each pixel value above 0 is considered as the label of a corresponding nucleus. We have annotated a considerable amount of cells, including normal epithelial and myoepithelial breast cells (localized in ducts and lobules), invasive carcinomatous cells, fibroblasts, endothelial cells, adipocytes, macrophages and inflammatory cells (lymphocytes and plasmocytes). In total, we have 33 images with a total of 2754 annotated cells, the maximum number of cells in one sample is 293 and the minimum number of cells in one sample is 5. We also have on average 83 cells per sample with a high standard deviation of 63. See figure 1 for some examples. The patients were randomly picked from an unpublished study on triple negative breast cancer.
We used ITK-snap for the annotation.
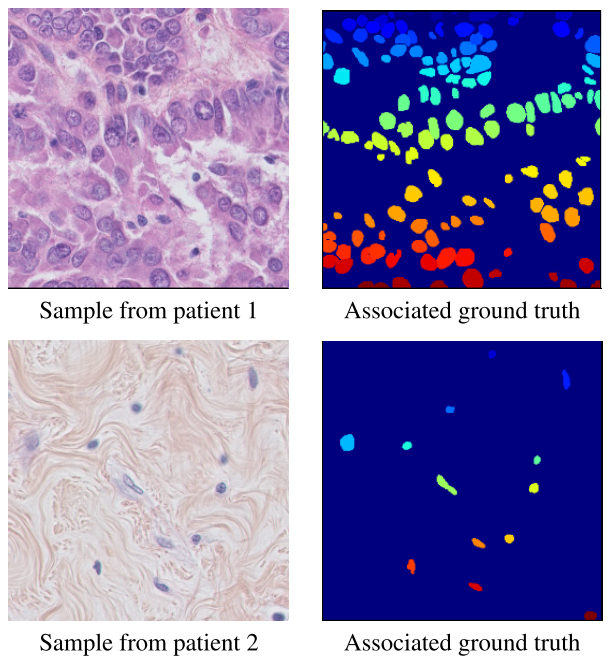 Figure 1: Random samples from the dataset
Figure 1: Random samples from the dataset
We propose a novel post-processing scheme for the probability map.
We benchmarked several deep learning algorithms on this dataset. See table 1 for metric comparison and figure 2 for a visual comparison of methods.
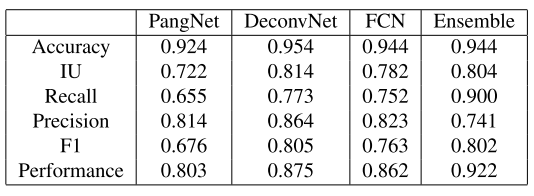 Table 1: Metric comparaison of methods
Table 1: Metric comparaison of methods
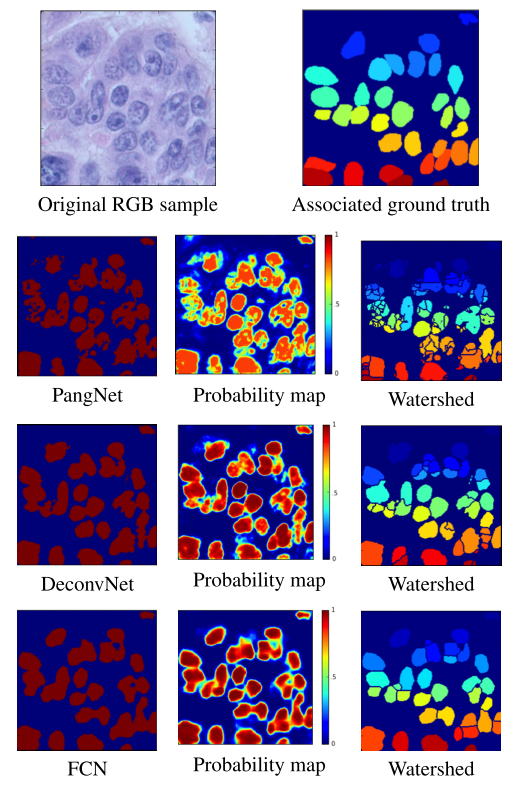 Figure 2: Visual comparaison of methods
Figure 2: Visual comparaison of methods
FCN: refers to the method in the paper: Long, Jonathan, Evan Shelhamer, and Trevor Darrell. “Fully convolutional networks for semantic segmentation.” Proceedings of the IEEE conference on computer vision and pattern recognition. 2015.
PangNet: refers to the method in the paper: Pang Baochuan, Zhang Yi, Chen Qianqing, Gao Zhifan, Peng Qinmu and You Xinge, “Cell nucleus segmentation in color histopathological imagery using convolutional networks,” in Pattern Recognition (CCPR), 2010 Chinese Conference on. IEEE, 2010, pp. 1–5.
DeconvNet: refers to the method in the paper: Hyeonwoo Noh, Seunghoon Hong, Bohyung Han, “Learning deconvolution network for semantic segmentation,” arXiv preprint arXiv :1505.04366, 2015.
ITK-snap: refers to the software developed in the paper: Paul A. Yushkevich, Joseph Piven, Heather Cody Hazlett, Rachel Gimpel Smith, Sean Ho, James C. Gee, and Guido Gerig, “User-guided 3D active contour segmentation of anatomical structures : Significantly improved efficiency and reliability,” Neuroimage, vol. 31, no. 3, pp. 1116–1128, 2006.
